Comprehensive Genomic Profiling

Detect multiple biomarkers in a single assay
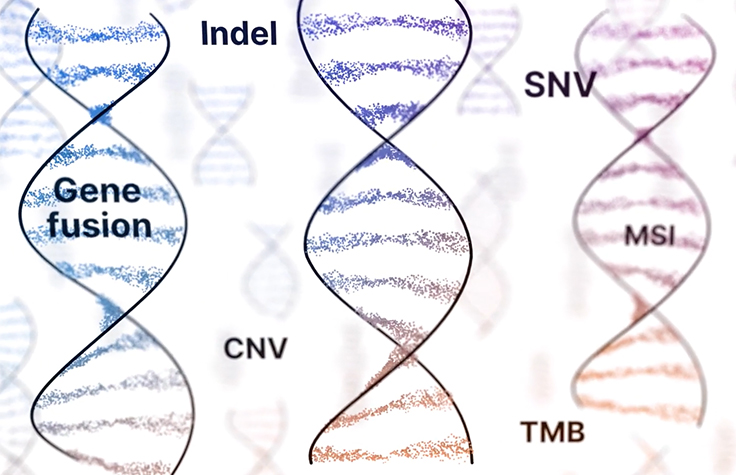
CGP can detect biomarkers at nucleotide-level resolution and typically comprises all major genomic variant classes (single nucleotide variants, indels, copy number variants, fusions, and splice variants). Additionally, CGP can detect genomic signatures such as TMB and MSI (tumor mutational burden and microsatellite instability, respectively), maximizing the ability to find clinically actionable alterations.

Consolidate testing to save time and precious samples
CGP consolidates biomarker detection into a single multiplex assay, eliminating the need for iterative testing. With a single test, you can simultaneously detect both common and rare biomarkers to increase the likelihood of identifying actionable alterations. This potentially provides faster results, limits the input of precious biopsy samples, and may reduce the risks and costs associated with rebiopsy.1,2

Identify actionable alterations
CGP can offer results for both actionable and potentially actionable biomarkers to help identify more effective therapeutic paths and innovative clinical trial options for cancer patients. When tissue biopsies are unavailable, CGP from liquid biopsy may provide helpful information about a tumor's genomic make-up. CGP using tissue and liquid biopsy together may reveal more insights into a tumor's composition.3,4
Percentage of potentially actionable alterations
Multiple studies have demonstrated the ability of CGP to identify potentially clinically relevant genomic alterations, across different tumor types.
| Potentially actionable variants identified in patient samples | Patient cohort |
|---|---|
 |
Single-center, prospective study with 339 patients. Refractory cancers, multiple types: ovarian (18%), breast (16%), sarcoma (13%), renal (7%), and others5 |
 |
Prospective study with 100 patients; diverse-histology, rare, or poor-prognosis cancers6 |
 |
Prospective study with 10,000 patients with advanced cancer across a vast array of solid tumor types7 |
 |
Retrospective study with 96 patients across multiple tumor types8 |
 |
6832 NSCLC patients9 |
The percent of actionable alterations identified in each study varies according to patient cohort, study type, CGP panel used, and criteria for categorizing a genomic alteration as actionable.
Data on file.

Improving patient outcomes with CGP
Discover how a single next-generation sequencing assay can reveal hundreds of actionable cancer biomarkers, reduce assay times, save valuable tissue samples, and transform lives. Whether you're a clinician or researcher, this guide can help you better understand cancer care with data-driven insights.
How CGP Compares to Other Sequencing Methods
CGP vs single-gene assays
Single-gene assays are limited to a single biomarker. Many times these assays do not cover the entire gene sequence, with the risk of missing important gene alterations.11
An iterative single-gene testing approach can lead to tissue depletion and repeat biopsies.11,13,14
CGP vs targeted panels
Targeted panels typically offer coverage of specific genes instead of the entire coding sequence. As a result, they can miss important alterations.7
A comprehensive single assay that assesses a wide range of biomarkers increases the chances of obtaining relevant information vs. targeted panels.
CGP vs exome sequencing
Not only can whole-exome sequencing be cost- prohibitive when developing personalized therapies, but it can also lead to inadequate coverage to detect important variants in lower frequencies due to the need for high amounts of sequencing.15-19
Reasons to bring CGP in-house
Integrating comprehensive genomic profiling into your lab’s in-house test menu can unlock a range of valuable advantages, including:

Delivering results faster (as compared to send-out services)

Reducing Quantity Not Sufficient (QNS) rates

Increasing number of informed cases
Building a database for future studies
Amplifying the role of the pathologists on the care team

Solutions for CGP
TruSight Oncology Comprehensive products are IVD test kits that enable genomic profiling based on DNA and RNA while consolidating multiple iterative tests into one.
Learn more about TruSight Oncology ComprehensivePeer-reviewed publications
Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer
Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial
Feasibility and utility of a panel testing for 114 cancer-associated genes: a hospital-based study
References
- Pennel AP, Mutebi A, Zheng-Yi Z, et al. Economic Impact of Next-Generation Sequencing Versus Single-Gene Testing to Detect Genomic Alterations in Metastatic Non–Small-Cell Lung Cancer Using a Decision Analytic Model. JCO Precis Oncol. 2019.
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn. 2018 Mar;20(2):129-159.
- Aggarwal C, Thompson JC, Black TA, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019 Feb 1;5(2):173-180.
- Tukachinsky H, Madison RW, Chung JH, et al. Genomic analysis of circulating tumor DNA in 3,334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms. Clin Cancer Res. 2021 Feb 8:clincanres.CCR-20-4805-E.2020.
- Wheler JJ, Janku F, Naing A et al. Cancer Therapy Directed by Comprehensive Genomic Profiling: A Single Center Study. Cancer Res. 2016 Jul 1;76(13):3690-701.
- Hirshfield KM, Tolkunov D, Zhong H. Clinical Actionability of Comprehensive Genomic Profiling for Management of Rare or Refractory Cancers. Oncologist. 2016 Nov;21(11):1315-1325.
- Zehir A, Benayed R, Shah R et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017 Jun;23(6):703-713.
- Reitsma M, Fox J, Borre PV, et al. Effect of a Collaboration Between a Health Plan, Oncology Practice, and Comprehensive Genomic Profiling Company from the Payer Perspective. Journal of Managed Care & Specialty Pharmacy. 2019 Jan 11:1-10
- Suh JH, Johnson A, Albacker L, et al. Comprehensive Genomic Profiling Facilitates Implementation of the National Comprehensive Cancer Network Guidelines for Lung Cancer Biomarker Testing and Identifies Patients Who May Benefit From Enrollment in Mechanism-Driven Clinical Trials. Oncologist. 2016 Jun;21(6):684-91.
- Kopetz S, Shaw K, Lee J, et al. Use of a Targeted Exome Next-Generation Sequencing Panel Offers Therapeutic Opportunity and Clinical Benefit in a Subset of Patients With Advanced Cancers. JCO Precision Oncology. 2019;3:1-14.
- Drilon A, Wang L, Arcila ME, et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin Cancer Res. 2015;21(16):3631-363.
- Ali SM, Hensing T, Schrock AB, et al. Comprehensive Genomic Profiling Identifies a Subset of Crizotinib-Responsive ALK-Rearranged Non-Small Cell Lung Cancer Not Detected by Fluorescence In Situ Hybridization. Oncologist. 2016 Jun;21(6):762-70.
- Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Annals of Oncology. 2015;26(7):1415-1421.
- Yu TM, Morrison C, Gold EJ, et al. Multiple Biomarker Testing Tissue Consumption and Completion Rates With Single-gene Tests and Investigational Use of Oncomine Dx Target Test for Advanced Non-Small-cell Lung Cancer: A Single-center Analysis. Clin Lung Cancer. 2018 Jan;20(1):20-29.e8.
- Buchhalter I, Rempel E, Endris V, et al. Size matters: Dissecting key parameters for panel-based tumor mutational burden analysis. Int J Cancer. 2019;144(4):848-858 .
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34.
- Pestinger V, Smith M, Sillo T, et al. Use of an integrated pan-cancer oncology enrichment NGS assay to Measure tumour mutational burden to detect clinically actionable variants. Mol Diagn Ther. 2020 Jun;24(3):339-349.
- Heydt C, Rehker J, Pappesch R, et al. Analysis of tumor mutational burden: correlation of five large gene panels with whole exome sequencing. Sci Rep. 2020 Jul 9;10(1):11387.
- Vanderwalde A, Spetzler D, Xiao N, et al. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018 Mar;7(3):746-756. Med. 2018 Mar;7(3):746-756.