Ready-to-Use Panels
Predesigned sequencing panels using AmpliSeq chemistry that target important genes associated with a disease or phenotype.
A comprehensive targeted resequencing solution offering ready-to-use and customizable panels for use with low-input DNA and RNA samples

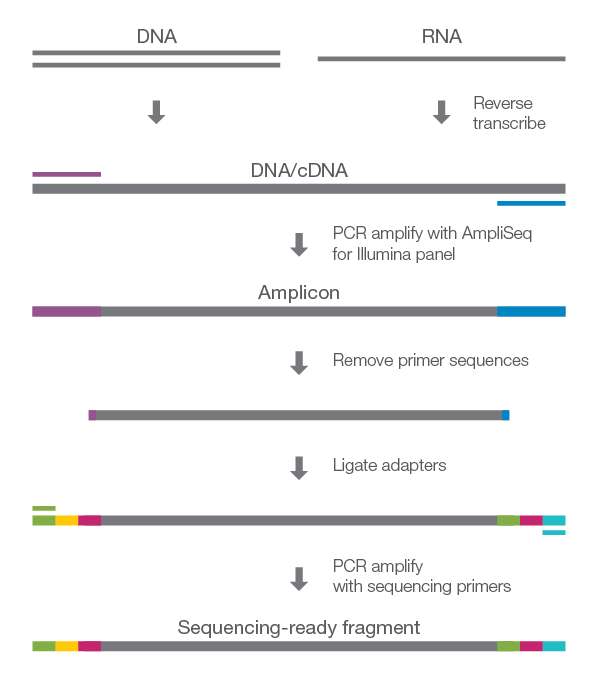
AmpliSeq for Illumina products deliver a fast, highly multiplexed PCR-based workflow for amplicon sequencing using low-input DNA and RNA samples. Compatible with Illumina next-generation sequencing (NGS) systems, AmpliSeq for Illumina chemistry offers high-quality data for disease research applications, enabling researchers to increase efficiency by targeting a few to hundreds of genes in a single run.
Predesigned sequencing panels using AmpliSeq chemistry that target important genes associated with a disease or phenotype.
Create custom sequencing panels optimized for content of interest with our free, user-friendly, online tool.
Create amplicon panels with the genes you want. Select from a catalog of pretested genes with known content relevant for inherited disease research.
These predesigned sequencing panels contain content selected and designed with input from leading disease researchers.
Targeted DNA and RNA research panel investigating 52 genes with known relevance to solid tumors.
FFPE-compatible panel for measuring T-cell diversity and clonal expansion in tumor samples by sequencing T-cell receptor beta chain rearrangements.
Targeted custom research panels optimized for sequencing specific targets or genomic content of interest.
Library prep begins with multiplexed PCR amplification of your genomic regions of interest with as little as 1 ng of DNA or cDNA input. After PCR, remaining primers are digested and the remaining amplicons used to prepare libraries for targeted sequencing on an Illumina NGS system.
Sequencing leverages widely adopted sequencing by synthesis (SBS) chemistry. Data are analyzed on the cloud with the DRAGEN Amplicon pipeline or on-instrument with Local Run Manager to get accurate results without extensive bioinformatics resources.
Learn about the robust performance of AmpliSeq for Illumina from a wide variety of sample types.
Prepare high-quality libraries quickly and simply using a multiplexed PCR-based workflow. Library prep takes ~5-7 hr total, with 1.5 hr hands-on time. Sequencing takes 17-32 hr, and data analysis time varies.
User-friendly assay design:
AmpliSeq for Illumina panel options:
Learn how to use BaseSpace Sequence Hub to cost-effectively manage, analyze, and share AmpliSeq for Illumina data in the cloud.
Watch an overview of how to perform on-premises amplicon analysis using Local Run Manager (LRM).
You can analyze AmpliSeq for Illumina data with user-friendly secondary analysis workflows in the cloud via the DRAGEN Amplicon pipeline or on-instrument via Local Run Manager. DRAGEN DNA Amplicon aligns reads against reference genomes and calls small variants. DRAGEN RNA Amplicon performs differential expression analysis and gene fusion calling. Tertiary analysis is available on Correlation Engine.
AmpliSeq for Illumina products are compatible with all Illumina sequencing systems. Users most often use benchtop sequencing systems.
If your genes of interest are not available as an AmpliSeq for Illumina Ready-to-Use Panel, you can use DesignStudio Assay Design Tool to design an AmpliSeq for Illumina Custom Panel. The DesignStudio tool is a free web-based assay design software that enables researchers to submit target regions of interest and receive personalized panel content customized for their study.
To transfer your existing workflows or start using Illumina technology for the first time, review this resource:
Transitioning to AmpliSeq for Illumina on the iSeq 100 System
You can also contact Illumina Technical Support.
Contact Illumina Technical Support if you need assistance with DesignStudio Assay Design Tool. For help with ordering, contact a sales representative.
Perform ultra-deep sequencing of PCR amplicons with cost-effective analysis of hundreds of target genomic regions in one assay.
Learn how NGS opens doors for clinical cancer research to help inform personalized medicine approaches.
Explore how NGS can efficiently identify causative variants associated with rare and inherited genetic disorders.
Contact an Illumina specialist to learn more about AmpliSeq for Illumina products.
Your email address is never shared with third parties.