ATAC-Seq for Chromatin Accessibility Analysis
ATAC-Seq (assay for transposase-accessible chromatin with sequencing)
A rapid, sensitive method for profiling accessible chromatin across the genome with high-throughput capabilities
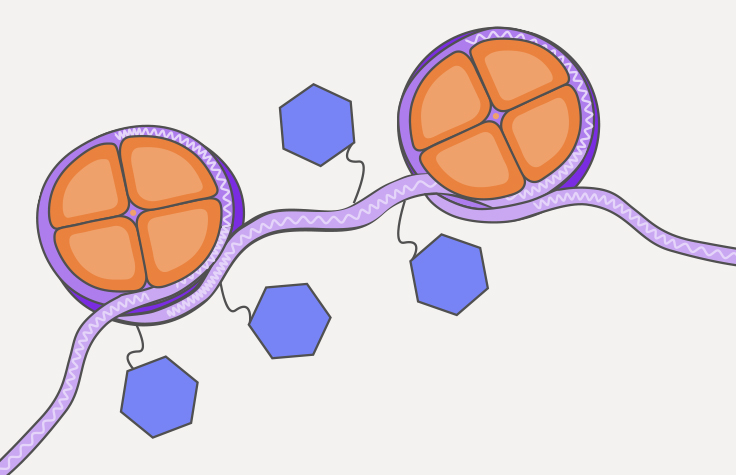
What is ATAC-Seq?
The assay for transposase-accessible chromatin with sequencing (ATAC-Seq) is a popular method for determining chromatin accessibility across the genome. By sequencing regions of open chromatin, ATAC-Seq can help you uncover how chromatin packaging and other factors affect gene expression.
ATAC-Seq does not require prior knowledge of regulatory elements, making it a powerful epigenetic discovery tool. It has been used to better understand chromatin accessibility, transcription factor binding, and gene regulation in complex diseases, embryonic development, T-cell activation, and cancer.1,2 ATAC-Seq can be performed on bulk cell populations or on single cells at high resolution.
How does ATAC-Seq work?
In ATAC-Seq, genomic DNA is exposed to Tn5, a highly active transposase. Tn5 simultaneously fragments DNA, preferentially inserts into open chromatin sites, and adds sequencing primers (a process known as tagmentation). The sequenced DNA identifies the open chromatin and data analysis can provide insight into gene regulation.
Additionally, ATAC-Seq can be combined with other methods, such as RNA sequencing, for a multiomic approach to studying gene expression.3,4 Subsequent experiments often include ChIP-Seq, Methyl-Seq, or Hi-C-Seq to further characterize forms of epigenetic regulation.
How to adapt ATAC-Seq for single cells
Dr William Greenleaf from Stanford University discusses ATAC-Seq development and its adaptation for single cells. In addition, Drs Bing Ren and Sebastian Preissl, experts in genomics and epigenetics, discuss single-cell ATAC-Seq using combinatorial indexing.
Watch webinar
Key applications of ATAC-Seq
Chromatin accessibility analysis with ATAC-Seq can provide valuable insights into the regulatory landscape of the genome. Popular applications include:
- Nucleosome mapping
- Transcription factor binding analysis
- Novel enhancer identification
- Exploration of disease-relevant regulatory mechanisms
- Cell type–specific regulation analysis
- Evolutionary studies
- Comparative epigenomics
- Biomarker discovery

NGS accelerates gene regulation and transcriptomics research
Learn how NGS is advancing gene expression and regulation research in various fields. See how gene regulation studies with methods such as ATAC-Seq and methylation sequencing can complement RNA sequencing.
Download eBookCoverage recommendations for ATAC-Seq
The minimum required sequencing coverage for ATAC-Seq varies according to research objectives. This table provides some guidelines for common applications.
We recommend using paired-end sequencing reads for ATAC-Seq. Compared to single-read sequencing, paired-end reads offer:
- Higher unique alignment rates
- Improved removal of PCR duplicates
- More complete information for accessible sequences
- Greater precision in categorizing reads as nucleosome-free, mono-nucleosomal, or di-nucleosomal
| Research goal | Recommended depth |
|---|---|
| Identification of open chromatin differences in human samples | ≥ 50M paired-end reads |
| Transcription factor footprinting to construct gene regulatory networks | > 200M paired-end reads |
| Single-cell analysis | 25K–50K paired-end reads per nucleus/cell |
As experimental needs can vary, we encourage you to consult the scientific literature to determine the right level of coverage for your project.
Publications using ATAC-Seq
Decoding gene regulation in human T cells
Read how scientists employ ATAC-Seq to identify regulators influencing chromatin states to control gene expression.
Improving metagenome assemblies using ATAC-Seq
Learn how investigators demonstrate the feasibility and benefits of ATAC-Seq to aid their studies on bacterial genomes.
Characterizing genomic architecture in cancer
See how scientists leverage ATAC-Seq to better understand genome topology in cancer using 15 primary human cancer types from The Cancer Genome Atlas.
Multiomics quick start: ATAC-Seq and RNA-Seq in single cells
In this video, learn how to perform ATAC-Seq and RNA-Seq in single cells to link mechanisms of gene regulation with phenotype. Illumina scientists describe best practices for sample prep, library prep, and sequencing to help you get started with your first single-cell multiomics project.
ATAC-Seq allows you to ask questions about the epigenetic variability in complex or rare tissues and epigenomic landscape in populations of cells that haven’t been observable at the genome-wide level before.
Additional resources
Profiling the transcriptome and epigenome
This technical note describes simultaneous profiling of the transcriptome and epigenome from single cells using 10x Genomics Chromium Single Cell Multiome ATAC + Gene Expression.
The regulatory landscape of A. thaliana
Join us for this webinar on single-cell ATAC-Seq of Arabidopsis thaliana. The speakers discuss an analytical framework to infer the regulatory networks that govern plant development.
Optimizing sample prep for single-cell applications
In this webinar, industry experts give insights into sample preparation techniques and best practices for ATAC-Seq as well as scRNA-Seq, sNuc-Seq, and other assays.
Cancer epigenetics
Explore how ATAC-Seq can be used to probe epigenetic features in cancer, along with guides, products, and other resources to help you get started.
RNA sequencing
RNA-Seq uses NGS to analyze expression across the transcriptome, allowing scientists to detect known or novel features and quantify RNA.
ATAC-Seq FAQ
References
- Cusanovich DA, Reddington JP, Garfield DA, et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature. 2018; 555:538-542. 13.
- Gate RE, Cheng CS, Aiden AP, et al. Genetic determinants of co-accessible chromatin regions in activated T-cells across humans. Nat Genet. 2018; 50:1140-1150.
- Luo L, Gribskov M, Wang S. Bibliometric review of ATAC-Seq and its application in gene expression. Briefings Bioinform. 2025;23:1-13. doi: 10.1093/bib/bbac061.
- Cao J, Cusanovich DA, Ramani V, et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380-1385.