
NovaSeq X Series ordering
Advanced chemistry, optics, and informatics combine to deliver exceptional sequencing speed and data quality, outstanding throughput, and scalability.
Comprehensive next-generation sequencing (NGS) assay that targets 170 genes associated with solid tumors.
Assay time
Hands-on time
Input quantity
The TruSight Tumor 170 bundle kits that contain NextSeq 500/550 v2 sequencing reagents (Catalog nos. OP-101-1003 and 20018621) have been discontinued. The TruSight Tumor 170 Kit with NextSeq v2.5 Reagents (Catalog no. 20028821) is the recommended replacement product. The TruSight Tumor 170 Kit Plus Watson for Genomics (Catalog no. 20018622) has also been discontinued. If you require that analysis, continue to work with IBM directly. Illumina remains committed to providing you with high-quality support and service.
TruSight Tumor 170 is an NGS assay that assesses 170 genes associated with common solid tumors.
Maximize single-assay efficiency using DNA and RNA from a single sample to assess multiple variant types, preserving valuable tissue, time, and resources. Expert-selected evidence-based content provides researchers with comprehensive coverage of variants most likely to play a role in tumorigenesis.
DNA and RNA libraries are prepared, sequenced, and analyzed simultaneously for efficient assessment of numerous types of somatic variants.
Use the TruSight Tumor 170 App on BaseSpace Sequence Hub for sequencing data analysis and management. Local secondary analysis is available on a Docker-based software.
| Assay time | ~2 days |
|---|---|
| Automation details | Explore available automation methods |
| Cancer type | Solid tumor |
| Content specifications | Probes enrich for full coding sequences of 170 genes. Calls single nucleotide variants, small insertions, and deletions in 151 genes, amplifications in 59 genes, and fusions plus splice variants in 55 genes. |
| Description | Perform comprehensive somatic variant detection research in solid tumors using variant calling information from both DNA and RNA. |
| Hands-on time | ~10.5 hr |
| Input quantity | 40 ng DNA and/or RNA |
| Instruments | NextSeq 550 System, NextSeq 500 System, HiSeq 2500 System |
| Method | Targeted DNA sequencing, Targeted RNA sequencing, Target enrichment |
| Multiplexing | Up to 32-plex for DNA using both index sets, up to 16-plex for RNA |
| Nucleic acid type | DNA, RNA |
| Specialized sample types | Low-input samples, FFPE tissue |
| Species category | Human |
| Technology | Sequencing |
| Variant class | Gene fusions, Somatic variants, Structural variants, Single nucleotide variants (SNVs), Insertions-deletions (indels), Copy number variants (CNVs) |
Requires use of NextSeq 500/550 v2.5 reagents and Pierian interpretation software.
TruSight Tumor 170 provides efficient comprehensive coverage of cancer-related variants (small variants, amplifications, splice variants, and fusions) in RNA and DNA obtained from a single FFPE sample.
| Instrument | Recommended number of samples | Read length |
|---|---|---|
| NextSeq 550 System | Samples per run (high output): 16 (8 DNA + 8 RNA), 10 if DNA only, 16 if RNA only |
2 × 101 bp (max recommended) |
Access a broad range of fixed and custom next-generation sequencing panels targeting known cancer-related gene variants.
Next-generation sequencing (NGS) provides a comprehensive method for assessing the majority of genes associated with solid tumors.
Illumina provides an innovative portfolio of NGS systems, products, and services for many phases of the drug development pipeline.
| TruSight Tumor 170 | TruSight Oncology 500 High-Throughput | TruSight Oncology 500 v2 | TruSight Oncology 500 ctDNA v2 | |
|---|---|---|---|---|
| Assay time | ~2 days | 4–5 days from sample to results | 3–4 days from sample input to final results | 2.5–4 days from purified nucleic acid to variant results |
| Automation details | Explore available automation methods | Explore available automation methods | Explore available automation methods | Explore available automation methods |
| Cancer type | Solid tumor | Pan-cancer, Solid tumor | Pan-cancer, Solid tumor | Pan-cancer, Solid tumor |
| Content specifications | Probes enrich for full coding sequences of 170 genes. Calls single nucleotide variants, small insertions, and deletions in 151 genes, amplifications in 59 genes, and fusions plus splice variants in 55 genes. | Targeted sequencing of DNA from 523 genes of interest and RNA from 55 genes, for a total of 1.94 Mb panel size. MSI and TMB measurement included. |
Targeted sequencing of DNA from 523 genes and RNA from 55 genes for a total of 1.94 Mb panel size. MSI and TMB measurement included. The included HRD panel † includes coverage of ~25K SNPs to assess homologous recombination deficiency through a comprehensive genomic instability score (LOH+TAI+LST) powered by Myriad Genetics. †TruSight Oncology 500 v2 is not available for sale in Japan |
Targeted sequencing of DNA for a total of 1.94 Mb panel size: 523 genes for small variants, 59 genes for CNVs, 23 genes for gene rearrangements, bMSI (> 2300 loci), bTMB (1.94 Mb). |
| Description | Perform comprehensive somatic variant detection research in solid tumors using variant calling information from both DNA and RNA. | A high-throughput comprehensive NGS assay to identify key biomarkers in guidelines and >1K clinical trials from a streamlined workflow using the NextSeq 1000 System, NextSeq 2000 System, NovaSeq 6000 System, NovaSeq 6000Dx instrument (in Research Mode), or NovaSeq X Series. Includes coverage of immuno-oncology biomarkers TMB and MSI. | Enables comprehensive genomic profiling of solid tumors using DNA and RNA from FFPE tissue. Provides variant detection and biomarker assessment from a single sample for broad tumor characterization. | Provides a noninvasive research method for comprehensive genomic profiling of liquid biopsy samples (ctDNA from blood plasma). This liquid biopsy approach provides insights about intra- and inter-tumor heterogeneity using a minimally invasive sample collection approach to complement tissue-based CGP. |
| Hands-on time | ~10.5 hr |
~2.5 hr for automated workflow ~10.5 hr for manual workflow |
~3.25 hrs for automated workflow ~5-7 hrs for manual workflow |
~1.5 hr for automated workflow ~2.5 hr for manual workflow |
| Input quantity | 40 ng DNA and/or RNA | 40 ng DNA and/or 40–80 ng RNA | 30 ng DNA (as low as 10 ng), 40 ng RNA (as low as 20 ng) | 20 ng cfDNA (4 ml of plasma) |
| Instruments | NextSeq 550 System, NextSeq 500 System, HiSeq 2500 System | NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NovaSeq 6000Dx in Research Mode, NovaSeq 6000 System, NovaSeq X Plus System | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NextSeq 550Dx in Research Mode, NovaSeq X System, NovaSeq 6000Dx in Research Mode, NovaSeq 6000 System, NovaSeq X Plus System | NextSeq 2000 System, NovaSeq X System, NovaSeq 6000Dx in Research Mode, NovaSeq 6000 System, NovaSeq X Plus System |
| Method | Targeted DNA sequencing, Targeted RNA sequencing, Target enrichment | Targeted DNA sequencing, Targeted RNA sequencing, Target enrichment | Targeted DNA sequencing, Targeted RNA sequencing, Target enrichment | Targeted DNA sequencing, Target enrichment |
| Multiplexing | Up to 32-plex for DNA using both index sets, up to 16-plex for RNA |
NextSeq 1000 and 2000: P2 flow cell 8 samples, P3 flow cell 24 samples, P4 flow cell 36 samples. NovaSeq 6000/Dx: SP flow cell 16 samples, S1 flow cell 32 samples, S2 flow cell 72 samples, S4 flow cell 192 samples. NovaSeq X Series*: 1.5B cartridge 32 samples, 10B cartridge 192 samples, 25B 480 samples. |
NextSeq 550/Dx: 8 samples/run. NexSeq 1000 and 2000: P2 flow cell 8 samples, P3 flow cell 24 samples, P4 flow cell 36 samples. NovaSeq 6000/Dx: SP flow cell 16 samples, S1 flow cell 32 samples, S2 flow cell 72 samples, S4 flow cell 192 samples. NovaSeq X/X+: 1.5 B 32 samples, 10B 192 samples, 25B 480 samples. |
NextSeq 2000 System: 4 samples on P4 flow cell. NovaSeq 6000 System: 4 samples on S1 flow cell, 8 samples on S2 flow cell, 24 samples on S4 flow cell. NovaSeq X Series: 4 samples on 1.5B flow cell, 24 samples on 10B flow cell, 64 samples on 25B flow cell. |
| Nucleic acid type | DNA, RNA | DNA, RNA | DNA, RNA | DNA |
| Specialized sample types | Low-input samples, FFPE tissue | FFPE tissue | FFPE tissue | Circulating tumor DNA, Blood |
| Species category | Human | Human | Human | Human |
| Technology | Sequencing | Sequencing | Sequencing | Sequencing |
| Variant class | Gene fusions, Somatic variants, Structural variants, Single nucleotide variants (SNVs), Insertions-deletions (indels), Copy number variants (CNVs) | Gene fusions, Somatic variants, Novel transcripts, Structural variants, Transcript variants, Single nucleotide variants (SNVs), Insertions-deletions (indels), Copy number variants (CNVs) | Gene fusions, Loss of heterozygosity (LOH), Somatic variants, Transcript variants, Single nucleotide variants (SNVs), Insertions-deletions (indels), Copy number variants (CNVs), Tumor mutational burden (TMB), Genomic instability score (GIS), Microsatellite instability (MSI), Novel transcripts, Single nucleotide polymorphisms (SNPs), Structural variants | Single nucleotide variants (SNVs), Insertions-deletions (indels), Copy number variants (CNVs), Blood microsatellite instability (bMSI), Blood tumor mutational burden (bTMB) |
Library Prep and Array Kit Selector
Find the right sequencing library preparation kit or microarray for your needs. Filter by method, species, and more. Compare, share, and order kits.
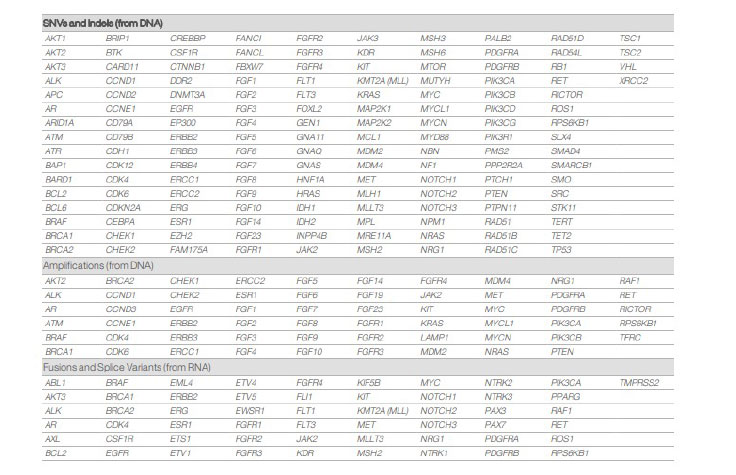

TruSight tumor 170 targets all coding exons per the current RefSeq database1 in 170 genes. The content includes 55 genes for fusions and splice variants, 148 SNVs and indels, and 59 amplifications.
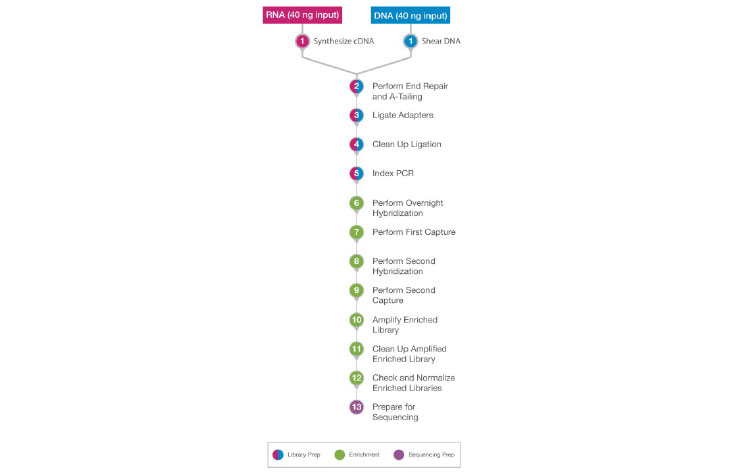

DNA and RNA samples follow the same workflow, after the cDNA synthesis step (for RNA) and the shearing step (for DNA).
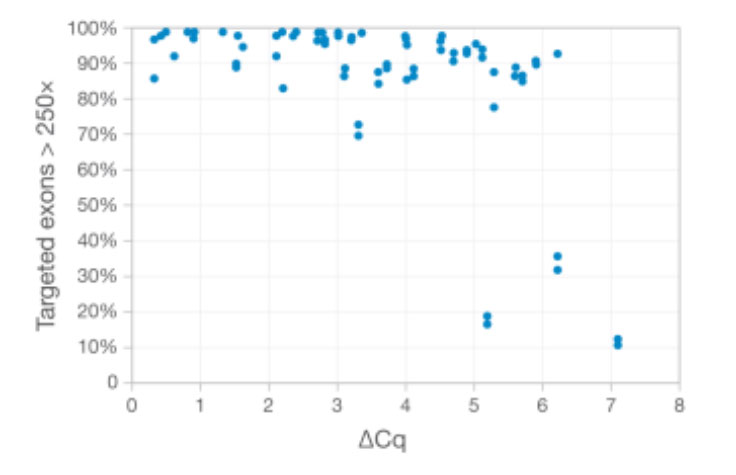

DNA from FFPE samples of varying quality was extracted and evaluated using TruSight Tumor 170 and sequenced on the NextSeq 500 System. Sample quality was assessed using qPCR to measure DNA amplification potential. The ΔCq value indicates the cycle threshold (Ct) value of each DNA sample minus the Ct value of a DNA standard.
Learn how a single assay interrogates both DNA and RNA to maximize the output from a single FFPE sample.
TruSight Tumor 170 Kit (24 Samples)
OP-101-1004
Includes library prep and enrichment reagents to allow for solid tumor profiling of 170 genes from FFPE samples.
List Price:
Discounts:
TruSight Tumor 170 Kit, With NextSeq v2.5 Reagents (24 Samples)
20028821
Includes library prep, enrichment, and sequencing consumables to allow for solid tumor profiling of 170 genes from FFPE samples.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
During library prep and enrichment, the DNA and RNA samples go through the same workflow following sample shearing (DNA) and cDNA synthesis (RNA). This results in the ability to process the DNA and RNA samples in parallel. The DNA and RNA libraries can then be sequenced on the same sequencing run.
Extraction reagents are not included in the TruSight Tumor 170 kit. The QIAGEN AllPrep DNA/RNA FFPE Kit (Catalog no. 80234) has demonstrated high yields of nucleic acids in comparison to other extraction methods for this assay. The QIAGEN AllPrep DNA/RNA FFPE Kit includes a DNase I digestion step during RNA extraction. You can select other commercially available extraction kits at your discretion.
TruSight Tumor 170 performance has been verified on the NextSeq 500 and NextSeq 550 Systems, and on the HiSeq 2500 System in rapid run mode.
Illumina recommends adding a minimum of 40 ng of DNA or RNA. For samples that are highly degraded, assay performance may be improved by using a maximum of 120 ng of DNA and 85 ng of RNA.
Coverage may vary based on sample quality and multiplexing level. Please review the DNA_SampleMetricsReport.txt and RNA_SampleMetricsReport.txt files produced by the TruSight Tumor 170 app for each analysis. This report contains a metric indicating the percent bases with > 100× coverage. A sequencing run with 8 high-quality DNA and 8 high-quality RNA samples (16 libraries) has demonstrated ≥ 99% bases with 100× coverage.
Use the TruSight Tumor 170 app available in BaseSpace Sequence Hub cloud environment or a local Docker App to analyze TruSight Tumor 170 libraries.
References:
1. O'Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733-45.
doi:10.1093/nar/gkv1189
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.