Voice of Customer: Illumina DNA Prep with Enrichment
Prof. Shinichi Yachida, So Takata (M.D.), and Yu Kanai (M.D.)
Graduate School of Medicine/Faculty of Medicine, Osaka University, Japan
Introduction
Whole-exome sequencing (WES) is widely used as a method for the comprehensive analysis of genomic aberrations of cancer cells and to study the evolution of cancer genomes.
In collaboration with Prof. Shinichi Yachida, So Takata, and Yu Kanai from the Graduate School of Medicine at Osaka University, formalin-fixed paraffin-embedded (FFPE) tumor tissue were subject to WES using Illumina DNA Prep with Enrichment (formerly Nextera™ Flex for Enrichment) to examine the ease of workflow and the quality of the sequencing libraries generated using FFPE samples.
Whole-exome Sequencing in Cancer Research
Prof. Yachida started utilizing WES when he was at the Department of Pathology at Johns Hopkins University. At the time, he performed WES using Sanger sequencing, and his research focused on evaluating clonal relationships between primary and metastatic tumors in pancreatic cancer patients. This research1 was published in Nature in 2011 and highlighted the importance of profiling genomic mutations in establishing the evolutionary history of cancer.
Currently, one area of Prof. Yachida’s research is focused on the genomic analysis of intractable and rare cancers. It is often challenging to obtain fresh-frozen tumor samples due to the scarcity of patients with rare cancers, thus FFPE samples are frequently used in these studies. Prof. Yachida pointed out the following four advantages of utilizing WES in his research:
Dr. Takata and Dr. Kanai
"The operation time is much shorter than that of the TruSeq Exome Kit, which I previously used. I think it is very convenient especially when there are many samples because it can process samples in half the time. I was also able to obtain the necessary data, and I think this is a useful kit."
– Dr. Takata
- The analysis pipeline is stable. WES has a long history and is widely used for the analysis of cancer genomes because it enables efficient search for somatic mutations in cancer.
- WES can be used for cell-free DNA. Only very small amounts of tumor DNA can be found in the blood, and deep sequencing is required for the accurate detection of mutations.
- WES is low cost. Many FFPE samples have low tumor cellularity, and these samples require a high read depth. WES is more cost-effective than whole-genome sequencing (WGS), and it enables sensitive mutation detection via deep sequencing.
- WES is better suited for exploratory research purposes than gene panels. For the identification of novel driver genes in rare cancers and specific diseases, WGS and WES are better suited to provide a comprehensive analysis. As most rare cancer samples are FFPE and are highly degraded with short fragments, WES is the preferred method.
"This was the first time that I used the sequencing library preparation kit, but the protocol is simple, and I could proceed with the procedure very efficiently. The quality of the obtained library was stable, and I think this is an easy-to-use product for everyone."
– Dr. Kanai
Prof. Yachida still has ambitious research aims utilizing WES. One example is his goal to perform WES to analyze the tumor microenvironment. This would require sequencing methods that include spatial information, and he looks forward to the development of WES techniques that would enable this.
Sequencing FFPE tumor samples using Illumina DNA Prep with Enrichment
Illumina DNA Prep with Enrichment is the fastest and most flexible targeted sequencing solution for DNA in the Illumina library prep portfolio. This method employs Illumina’s Bead-linked transposome (BLT) chemistry, integrating DNA extraction, fragmentation, library preparation, and library normalization steps. This helps to simplify the workflow, reduces the number of workflow steps and both hands-on and turnaround time. The entire workflow only takes around 6.5 hours with 2 hours of hands-on time, and there is no need for specialized equipment such as an ultrasonic homogenizer for DNA fragmentation.
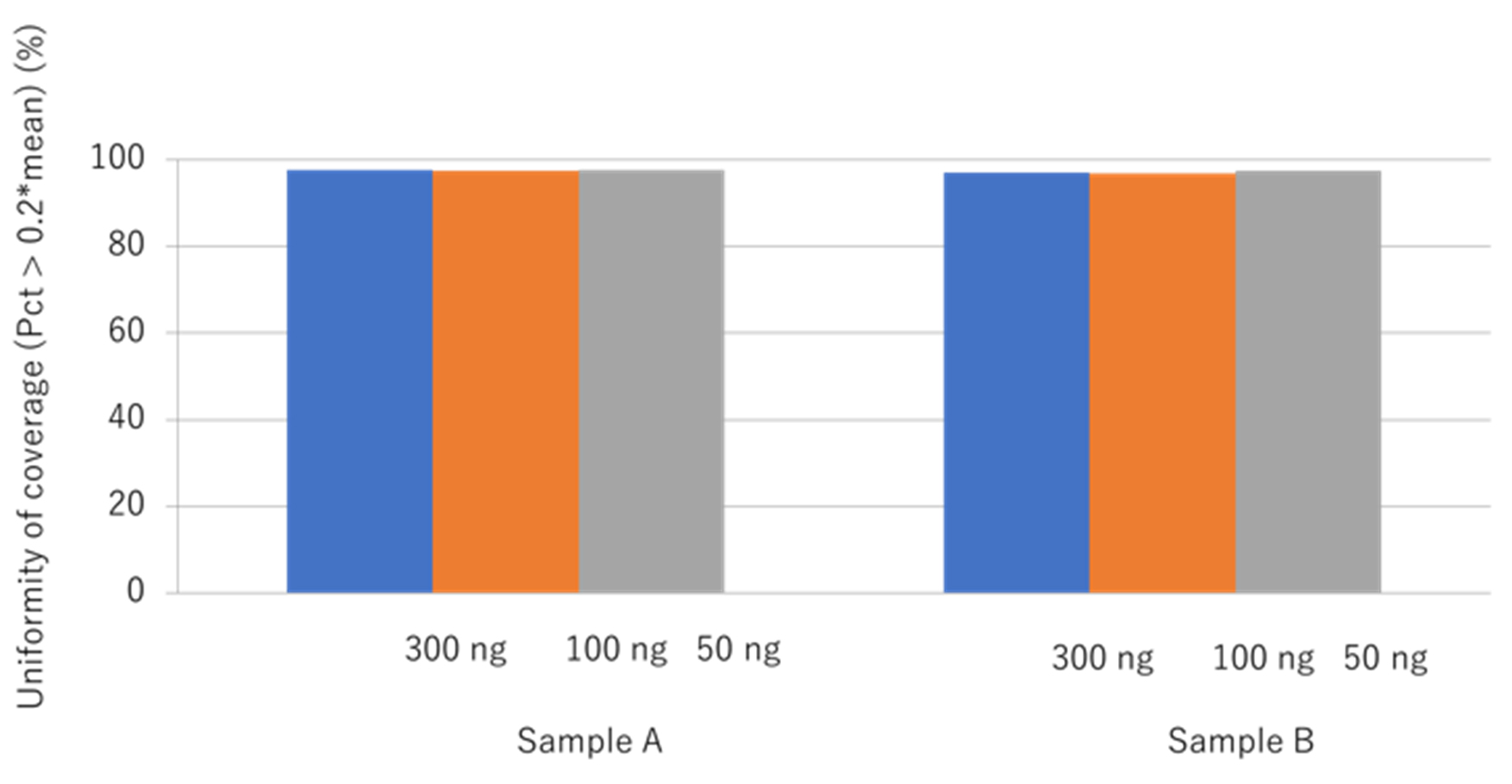
Illumina DNA Prep with Enrichment can support FFPE sample-derived DNA input amounts from 50 to 1,000 ng. To validate the impact of the DNA input amount on coverage uniformity, two different FFPE samples were divided into amounts of 50 ng, 100 ng, and 300 ng, respectively. The coverage uniformity was high for all samples, regardless of the quantity of DNA input (see Figure 1).
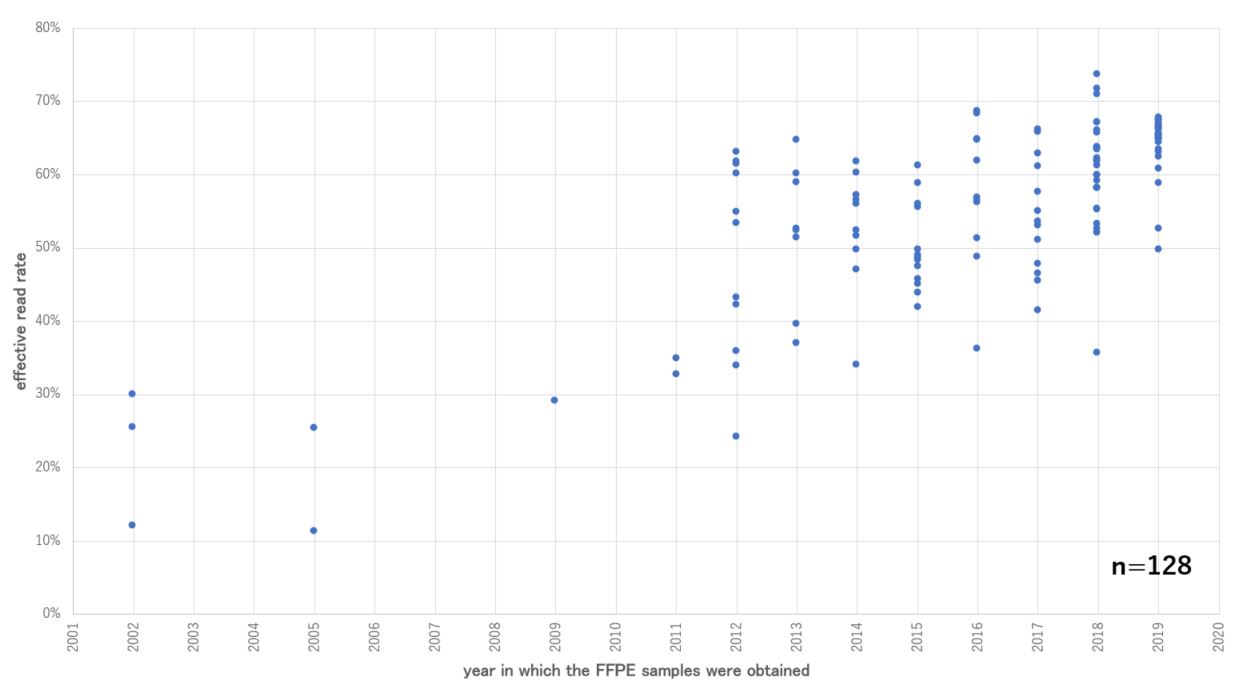
Illumina DNA Prep with Enrichment has robust performance even with older FFPE samples. Although the effective read rate2 of old FFPE samples is generally low, there was no decrease in the effective read rate observed, and relatively stable results were obtained even for FFPE samples from 2012 (Figure 2).
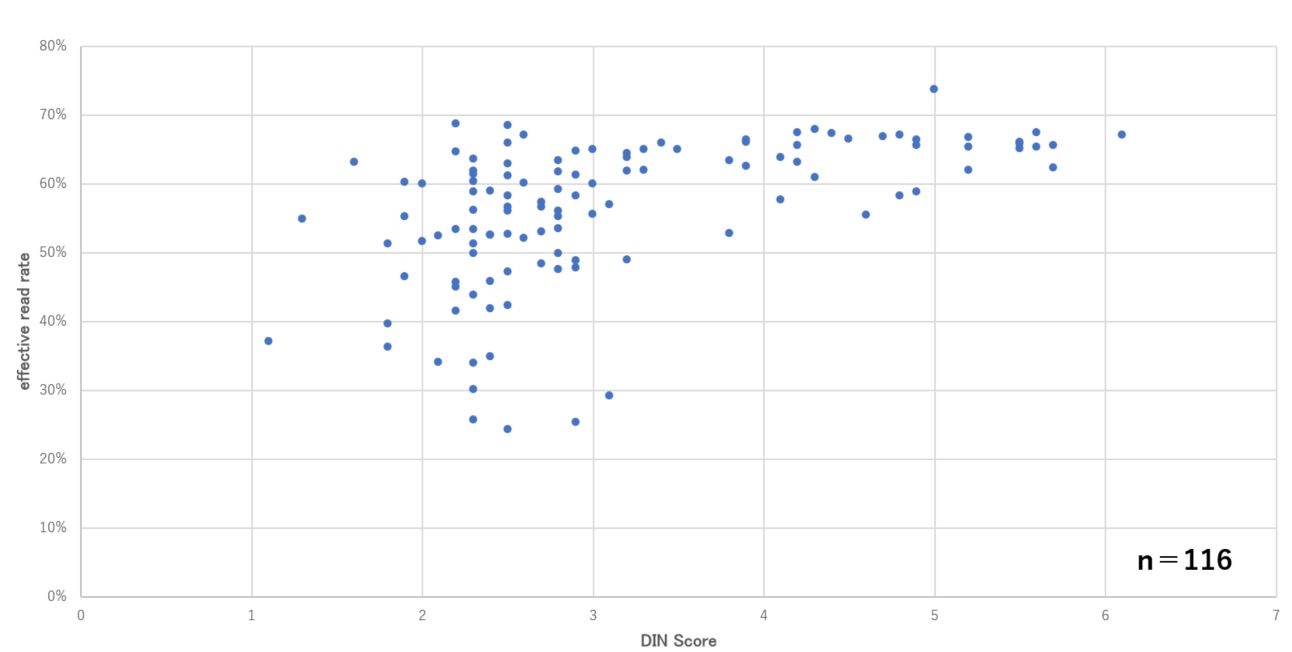
Looking at the DIN3 value, it was observed that a stable and effective read rate was obtained when the DIN value was 3 or higher, demonstrating the utility of the DIN value as a QC measure for obtaining good quality sequencing data (Figure 3).
Summary
Whole-exome sequencing with FFPE samples is an important application for cancer genome analysis. Illumina DNA Prep with Enrichment provides a rapid workflow with high quality sequencing data, even in FFPE tumor samples of varying quality and input amount.
References:
- Yachida, S., Jones, S., Bozic, I. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010). https://pubmed.ncbi.nlm.nih.gov/20981102/
- Effective read rate: Percentage of reads that can finally be used for analysis after duplicate reads, etc., have been removed from all reads.
- DIN stands for “DNA Integrity Number.” This is an index for objectively assessing the extent of genomic DNA degradation based on the profile that is obtained from Agilent Technologies, Inc.’s TapeStation System. The extent of genomic DNA degradation is indicated with a DIN value between 1 and 10 (1 = completely degraded state, 10 = almost undegraded state).