Whole-transcriptome profiling of large tissue areas
Whole-transcriptome profiling of large tissue areas
Dr Michal Lipinski of the Broad Institute shares how Illumina spatial technology enables cell type clustering and gene marker identification.
Enabling high-resolution, single cell–level whole-transcriptome analysis on Illumina high-throughput sequencing systems

Illumina spatial transcriptomics enables high-resolution, sequencing-based whole-transcriptome analysis in intact tissue sections. The technology supports hypothesis-free gene expression profiling while preserving spatial context, addressing limitations of imaging-based approaches, which are often constrained by predefined gene panels and limited tissue coverage.
Illumina advanced spatial technology, available in mid-2026, enables researchers to access broad transcriptomic profiles alongside localized gene expression patterns at single-cell resolution. This capability provides new opportunities to study spatially defined cell states, tissue architecture, and biological pathways across diverse research applications.

Illumina spatial technology helps researchers overcome the common limitations of existing platforms by resolving the tradeoff between high spatial resolution and whole-transcriptome depth. By enabling detailed mapping across large tissue areas without compromising sensitivity or scalability, the technology delivers clearer insights into cellular behavior and tissue organization.
Reveals cellular behavior within the spatial context of complex tissues
Supports unbiased, whole-transcriptome profiling in intact tissue sections
Preserves spatial heterogeneity by capturing large tissue regions
Offers streamlined analysis with built-in tools for segmentation and spatial visualization
Provides high spatial resolution combined with whole-transcriptome breadth
Delivers high sensitivity, enabling improved clustering to help identify distinct or rare cell types
Illumina spatial technology captures polyadenylated RNA transcripts on a substrate adapted from Illumina sequencing flow cell technology. This polyA-based design enables hypothesis-free, whole-transcriptome analysis with high spatial resolution across large tissue areas and is compatible with any eukaryotic species. The slide capture surface supports strong transcript recovery while preserving spatial fidelity within intact tissue sections.
The workflow begins with cryosectioning, H&E staining, and brightfield imaging to create a spatial reference. RNA is captured on barcoded slides and sequenced on high-throughput systems like the NovaSeq X Series, then combined with spatial barcodes and tissue images for segmentation, clustering, and spatial mapping. This enables high-resolution spatial analysis across applications ranging from pilot studies to large-scale research projects.
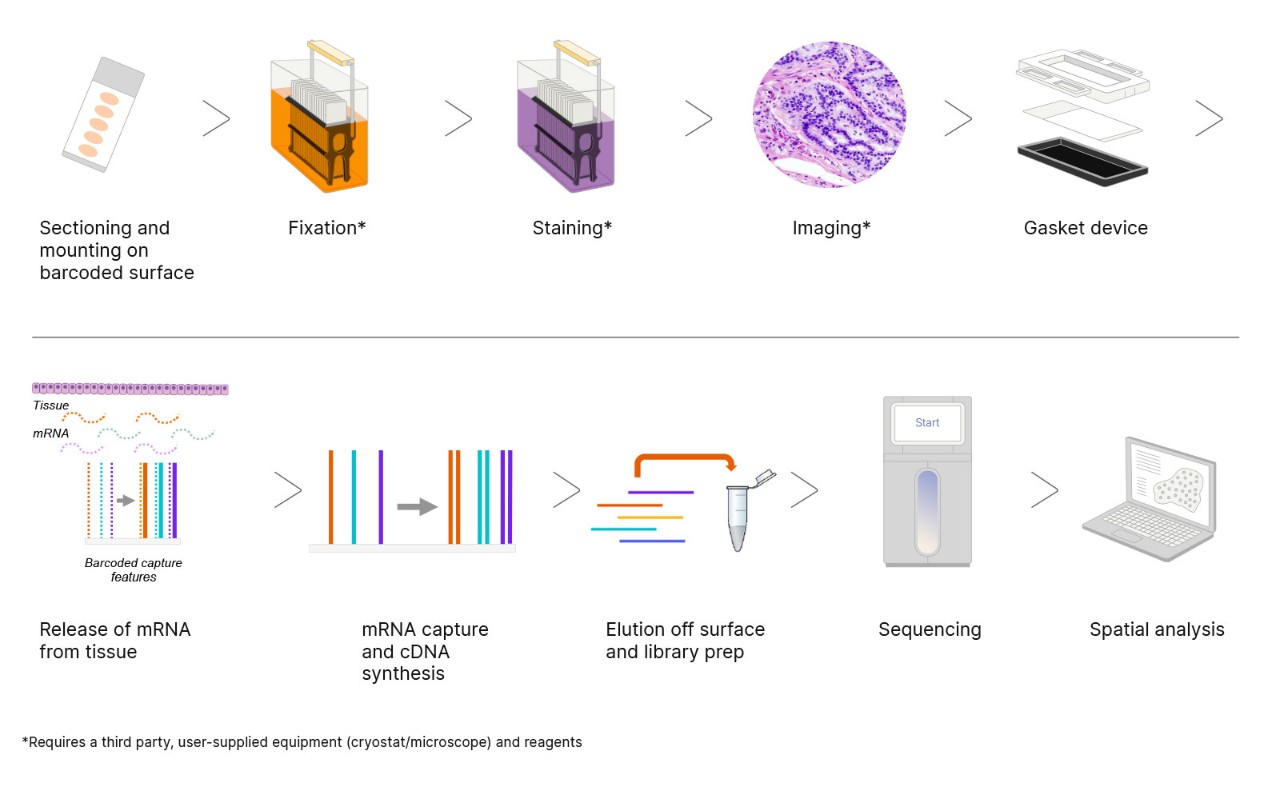
Figure 1: Overview of the Illumina spatial transcriptomics workflow, from tissue sectioning through spatial analysis.
Learn how to enable broad spatial profiling while balancing resolution and sensitivity. Navigate workflow options, explore resources for study planning, and more.
Your email address is never shared with third parties.
Whole-transcriptome profiling of large tissue areas
Dr Michal Lipinski of the Broad Institute shares how Illumina spatial technology enables cell type clustering and gene marker identification.
A spatial map of pregnant mouse brains
Dr Darren Segale, Sr Director of Scientific Research at Illumina, details how Illumina spatial technology revealed neuronal differences between virgin and pregnant mice at single-cell resolution with whole-transcriptome coverage.
Spatial sequencing reveals tumor environment changes
Dr Jasmine Plummer of St. Jude Children’s Research Hospital explains how high-resolution spatial sequencing aids prostate cancer research.
The analysis workflow integrates high-resolution tissue images, spatial barcodes, and gene expression profiles to map transcriptional activity in spatial context. Raw sequencing data are processed using the DRAGEN spatial pipeline, which aligns reads, deduplicates molecular identifiers, and generates spatial feature-barcode matrices. Tissue images and barcodes are also processed through this pipeline, with results visualized in Illumina Connected Multiomics. The workflow supports segmentation, clustering, and spatial mapping from raw imaging through transcriptomic analysis.
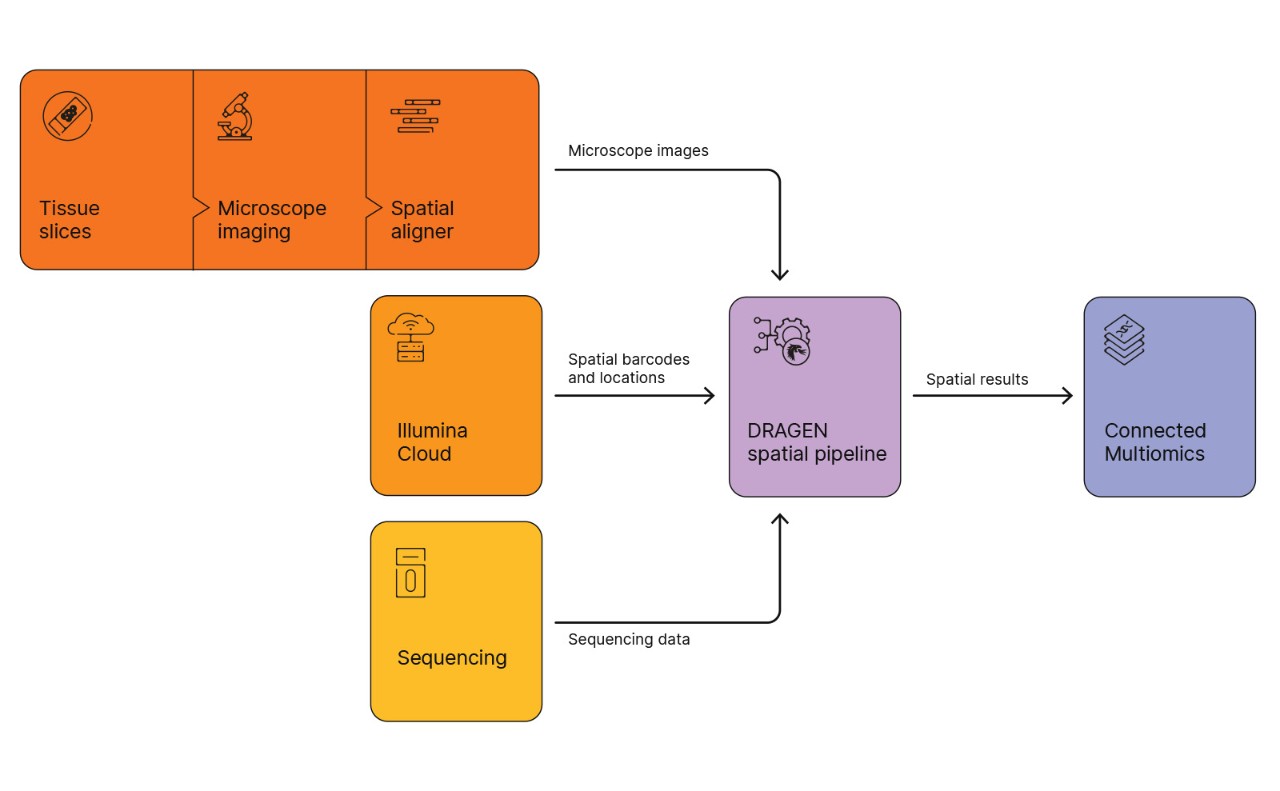
Figure 2: lIlumina spatial transcriptomics software workflow. Tissue images and spatial barcodes are processed through the DRAGEN spatial pipeline and visualized in Illumina Connected Multiomics.
Illumina Connected Multiomics, with DRAGEN Spatial Transcriptome, offers powerful, streamlined multiomic visualization and insights for spatial transcriptomics studies. Easily view spatial data overlaid on tissue images and perform additional tertiary analysis such as differential expression, marker gene identification, and cell typing.

Illumina spatial technology uses a large, advanced tissue placement area with continuous 1-µm surface features. The captured transcripts are binned with integrated cell segmentation through the Illumina spatial analysis pipeline to enable cell-level expression mapping.
Read the press release to learn more about Illumina spatial technology.
During development, we have seen that coverage varies based on tissue type, tissue quality, and sequencing depth. We have observed that the technology can detect ~2× more genes in a tissue section compared to other spatial solutions.1
The initial version of Illumina spatial technology is optimized for use with fresh-frozen tissue. A version supporting formalin-fixed, paraffin-embedded (FFPE) tissue is in development.
Due to the required depth of sequencing, we recommend using the NovaSeq X Series or NovaSeq 6000 System. The NextSeq 2000 System may be used for smaller tissue sections or tissue arrays.
Two slide formats will be available for spatial transcriptomic analysis: a large-format slide with a 50 x 15 mm capture area, and a second format with a smaller capture area (dimensions to be determined). Both versions will use a standard 75 × 25 mm microscope slide.
Cell atlasing human intestine using Illumina spatial solution
Dr Chenchen Zhu, Research Scientist at Stanford University, presents how Illumina spatial technology enables high-resolution cell atlasing of the human intestine.
Alveolar dysregulation in pulmonary fibrosis
Dr Nicholas Banovich, Associate Director of the Division of Bioinnovation and Genome Sciences at TGen, explains how Illumina spatial technology aids research in pulmonary fibrosis and epithelial remodeling.

Whole-transciptome spatial technology workflow
An ex-situ whole-transcriptome workflow providing large area capture substrate and scalable end-to-end software solution.
Poster
Map transcriptional activity within structurally intact tissue to unravel complex biological interactions using spatial RNA-Seq.
Study cellular differences often masked by bulk sampling, and explore high- and low-throughput single-cell RNA sequencing methods.
Simultaneously quantify cell surface protein and transcriptomic data within a single cell readout.
Interested in learning more about Illumina spatial technology? Sign up to receive updates.
Your email address is never shared with third parties.
References