
NextSeq 1000 and NextSeq 2000 System ordering
These sequencing systems offer expansive application breadth, operational simplicity, high flexibility and scalability, and proven performance.
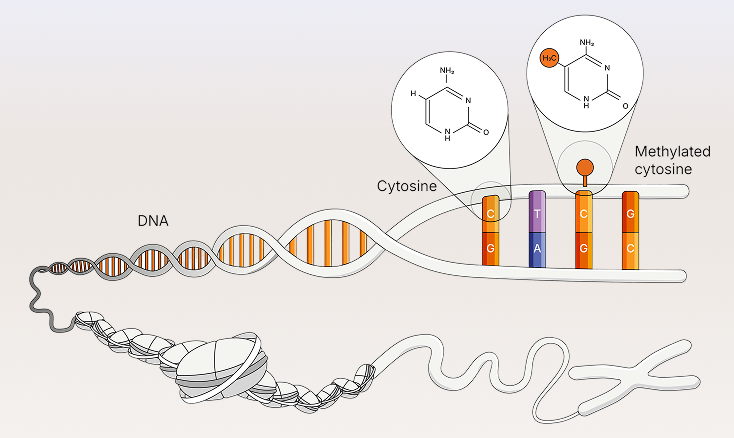
A single assay for targeted detection of five DNA bases (A, T, G, C, and 5mC), providing dual insights into genomic variants and methylation events.
Assay time
Input quantity
Limited time offer: 25% off first kit with promo code ILMN5B25
Now through May 2026, receive 25% off your first library prep kit
Limited quantity: 1
Illumina 5-Base DNA Prep with Enrichment offers a highly sensitive approach for detecting both DNA variants and methylation changes from a single sample preparation and sequencing run. The two-for-one targeted assay uses a cost-effective, library preparation-to-analysis workflow.
Detect DNA variants and methylation changes in one assay with an integrated workflow from library prep to analysis—no sample splitting required.
Simultaneously detect low-frequency variants and methylation from low-input samples. Alternatively, reduce sequencing costs by focusing coverage on regions of interest.
Create tailored dual-omic panels for Illumina 5-Base DNA Prep with Enrichment using the intuitive DesignStudio platform.
| Assay time |
< 3 days DNA to data Library prep time* < 11 hours * Based on processing 24 samples manually; includes DNA shearing (if applicable), and library quantification. Normalization and pooling time not included. |
|---|---|
| Automation capability | Liquid handling robot(s) |
| Automation details | Automation protocols are in development and will be added to the automation page when available. |
| Content specifications | Custom panels |
| Description | A streamlined assay for targeted detection of genomic variants and methylation events from the same sequencing reads. High-quality data is enabled by a novel enzymatic method and optimized analysis. Focus on specific targets of interest using hybrid-capture enrichment and custom-designed panels. |
| Input quantity |
1–20 ng cell-free DNA 50–100 ng genomic DNA |
| Instruments | NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NovaSeq 6000Dx in Research Mode, NovaSeq 6000 System, NovaSeq X Plus System |
| Mechanism of action | Enzymatic conversion of 5-methylcytosine to thymine, ligation-based addition of adapters and indexes, mechanical shearing of genomic DNA, hybridization-based target capture |
| Method | Custom sequencing, Methylation sequencing, Target enrichment |
| Multiplexing | Up to 384-plex indexing. 4-plex enrichment |
| Nucleic acid type | DNA |
| Specialized sample types | Blood, Low-input samples, Cell-free DNA |
| Species category | Any species, Human |
| Species details |
Supports custom panel design for a wide variety of species. Users can upload custom reference genome to run Illumina 5-base assay/analysis. Not all species have been tested and specifications are only applicable to human samples. |
| Technology | Sequencing |
| Variant class | Somatic variants, Differentially methylated cytosines, Germline variants, Single nucleotide variants (SNVs), Insertions-deletions (indels), Copy number variants (CNVs) |
Illumina 5-Base DNA Prep with Enrichment requires the use of DRAGEN secondary analysis. DRAGEN analysis can be accessed via a local DRAGEN server or the cloud through Illumina Connected Analytics or BaseSpace Sequence Hub. Index adapters are sold separately through Illumina.
Illumina 5-Base DNA Prep with Enrichment enables high-sensitivity, dual detection of DNA variants and methylation from low-input samples in a single, customizable assay while delivering precise 5-base insights with an efficient, cost-effective workflow from library prep to analysis.
Illumina 5-Base DNA Prep with Enrichment
Genome and methylome sequencing
Achieve simultaneous high-accuracy genome and methylome discovery in a single readout with the Illumina 5-base genome solution.
Explore the benefits of next-generation sequencing and microarrays for analyzing altered methylation patterns and other epigenetic changes in cancer.
Illumina offers solutions for epigenetic analysis. Use our tools to study epigenetic modifications and their impact on gene regulation.
| Illumina 5-Base DNA Prep with Enrichment | Illumina Cell-Free DNA Prep with Enrichment | Illumina DNA Prep with Enrichment | Infinium HTS iSelect Methyl Custom BeadChip | |
|---|---|---|---|---|
| Assay time |
< 3 days DNA to data Library prep time* < 11 hours * Based on processing 24 samples manually; includes DNA shearing (if applicable), and library quantification. Normalization and pooling time not included. |
~8.5–9.5 hr | ~6.5 hr | |
| Automation capability | Liquid handling robot(s) | Liquid handling robot(s) | Automated array loader | |
| Automation details | Automation protocols are in development and will be added to the automation page when available. | Explore available automation methods | ||
| Content specifications | Custom panels |
Custom: 0.5 - 15 Mb genomic content of interest, with Illumina Custom Enrichment Panels and Illumina Custom Enrichment Panel v2 Fixed panels: Content varies by panel. |
||
| Description | A streamlined assay for targeted detection of genomic variants and methylation events from the same sequencing reads. High-quality data is enabled by a novel enzymatic method and optimized analysis. Focus on specific targets of interest using hybrid-capture enrichment and custom-designed panels. | A fast, flexible, and scalable cfDNA library preparation kit for highly accurate mutation detection | Prepare sequencing libraries for a variety of experiments, including whole-exome, custom, and fixed panel targeted sequencing, from low DNA inputs and a variety of sample types in ~6.5 hours. | Using the DesignStudio Assay Design Tool, this product allows labs to create high-throughput, custom, 24- or 48-sample BeadChips to efficiently and accurately assess methylation for up to 100,000 CpG sites in human and select nonhuman samples. |
| Input quantity |
1–20 ng cell-free DNA 50–100 ng genomic DNA |
10–30 ng cfDNA | 10-1000 ng high-quality genomic DNA or 50-1000 ng FFPE DNA. (For blood and saliva, see the reference guide). | 250 ng DNA |
| Instruments | NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NovaSeq 6000Dx in Research Mode, NovaSeq 6000 System, NovaSeq X Plus System | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NextSeq 550Dx in Research Mode, NovaSeq X System, NovaSeq 6000Dx in Research Mode, NovaSeq 6000 System | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, MiSeqDx in Research Mode, MiniSeq System, NextSeq 550Dx in Research Mode, NovaSeq X System, NovaSeq 6000Dx in Research Mode, NovaSeq 6000 System, NovaSeq X Plus System, MiSeq i100 Plus System, MiSeq i100 System | iScan System |
| Mechanism of action | Enzymatic conversion of 5-methylcytosine to thymine, ligation-based addition of adapters and indexes, mechanical shearing of genomic DNA, hybridization-based target capture | Bead-bound transposomes and hybrid-capture chemistry | ||
| Method | Custom sequencing, Methylation sequencing, Target enrichment | Targeted DNA sequencing, Custom sequencing, Target enrichment | Targeted DNA sequencing, Custom sequencing, Exome sequencing, Target enrichment | Methylation array |
| Multiplexing | Up to 384-plex indexing. 4-plex enrichment | Up to 384 available unique dual indexes. Pre-enrichment pooling of up to 12-plex is tested. | ||
| Nucleic acid type | DNA | DNA | DNA | DNA |
| Specialized sample types | Blood, Low-input samples, Cell-free DNA | Cell-free DNA | Blood, Low-input samples, FFPE tissue, Saliva | Blood, Cell-free DNA, Saliva |
| Species category | Any species, Human | Human | Other, Human | Mouse, Human |
| Species details |
Supports custom panel design for a wide variety of species. Users can upload custom reference genome to run Illumina 5-base assay/analysis. Not all species have been tested and specifications are only applicable to human samples. |
Select non-human species available on DesignStudio only | ||
| Technology | Sequencing | Sequencing | Sequencing | Microarray |
| Variant class | Somatic variants, Differentially methylated cytosines, Germline variants, Single nucleotide variants (SNVs), Insertions-deletions (indels), Copy number variants (CNVs) | Gene fusions, Somatic variants, Single nucleotide variants (SNVs), Insertions-deletions (indels), Copy number variants (CNVs) | Single nucleotide polymorphisms (SNPs), Loss of heterozygosity (LOH), Somatic variants, Germline variants, Insertions-deletions (indels), Copy number variants (CNVs) | Differentially methylated cytosines |
Library Prep and Array Kit Selector
Find the right sequencing library preparation kit or microarray for your needs. Filter by method, species, and more. Compare, share, and order kits.
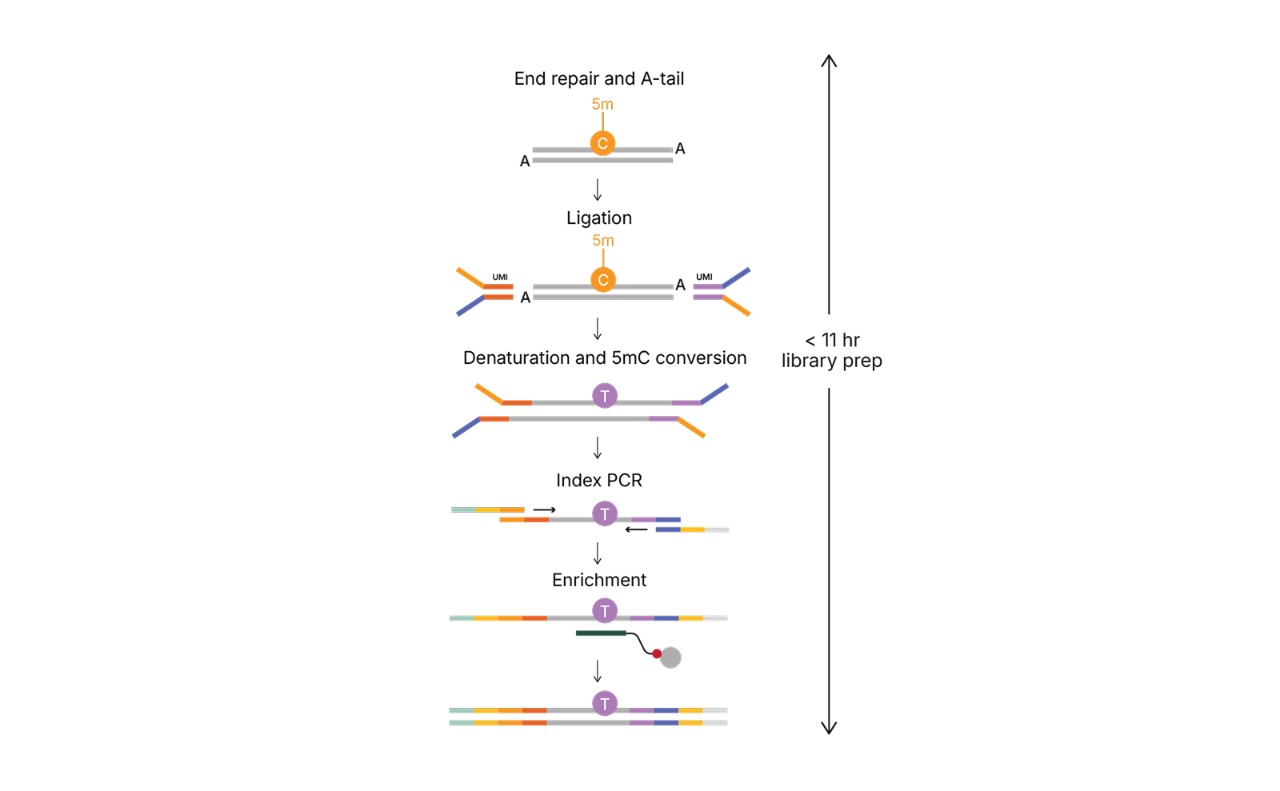

Library preparation steps for Illumina 5-Base DNA Prep with Enrichment
Optimized ligation-based library preparation includes a simple, one-step 5mC-to-T base conversion, requires minimal touchpoints, and is completed in fewer than 11 hours. Time includes DNA shearing (if applicable) and library quantification. Normalization and pooling times are not included.


Novel chemistry directly converts 5mC to T in a single enzymatic step
(A) Illumina 5-Base DNA Prep with Enrichment uses a one-step enzymatic process to convert only 5mC to T, yielding greater nucleotide diversity than (B) traditional bisulfite sequencing or on-market enzymatic methylation sequencing (EM-Seq), in which unmethylated C are converted to T.

Illumina 5-base solution: A comprehensive Workflow Guide
Discover the benefits of high-accuracy dual-omic data with DRAGEN 5-base pipelines and gain insights into the reagents, equipment, and user-supplied consumables required for successful implementation.

Decode DNA and methylation together with the Illumina 5-base solution
Join Fiona Kaper, VP and Head of Assay R&D at Illumina, as she discusses the innovative 5-base solution that simplifies and scales multiomic analysis. Discover how Illumina 5-base chemistry enables dual genomic and epigenomic insights in one assay, providing genetic variant and DNA methylation data with exceptional accuracy, simplicity, and scale.
An introduction to the Illumina 5-base solution
A streamlined, end-to-end NGS library prep and analysis workflow for simultaneous methylation profiling and high-accuracy genetic variant calling.
Illumina® 5-Base DNA Prep with Enrichment (24 Samples)
20140366
Includes library prep, enrichment and methylation detection reagents for preparing 24 libraries (6, 4-plex enrichment reactions) manually. Analyze data on the cloud using DRAGEN apps on Illumina Connected Analytics or BaseSpace Sequence Hub or on premise with DRAGEN Server. Perform data visualization using Illumina connected Multiomics (option). Design panel through DesignStudio. Purchase indexes and enrichment probe panel separately.
List Price:
Discounts:
Illumina Custom Enrichment Panel V2 (32 µl, 120 bp)
20073953
Illumina Custom Enrichment Panel V2 enables 120 bp double-stranded custom panel design from 100 up to 1,000,000 probes using DesignStudio or Concierge Services and provides 32 µl of material (sufficient for 8 enrichment reactions when paired with Illumina DNA Prep with Enrichment). Library prep, enrichment, and index adapter reagents need to be ordered separately.
Illumina Custom Enrichment Panel V2 (384 µl, 120 bp)
20073952
Illumina Custom Enrichment Panel V2 enables 120 bp double-stranded custom panel design from 100 up to 1,000,000 probes using DesignStudio or Concierge Services and provides 384 µl of material (sufficient for 96 enrichment reactions when paired with Illumina DNA Prep with Enrichment). Library prep, enrichment, and index adapter reagents need to be ordered separately.
Illumina Custom Enrichment Panel V2 (1536 µl, 120 bp)
20111339
Illumina Custom Enrichment Panel V2 enables 120 bp double-stranded custom panel design from 100 up to 1,000,000 probes using DesignStudio or Concierge Services and provides 1536 µl of material (sufficient for 384 enrichment reactions when paired with Illumina DNA Prep with Enrichment). Library prep, enrichment, and index adapter reagents need to be ordered separately.
Illumina® Unique Dual Indexes, LT (48 Indexes, 48 Samples)
20098166
Illumina® Unique Dual Indexes, LT (48 Indexes, 48 Samples)
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set A, Tagmentation (96 Indexes, 96 Samples)
20091654
Illumina® DNA/RNA UD Indexes Set A, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set B, Tagmentation (96 Indexes, 96 Samples)
20091656
Illumina® DNA/RNA UD Indexes Set B, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set C, Tagmentation (96 Indexes, 96 Samples)
20091658
Illumina® DNA/RNA UD Indexes Set C, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Illumina® DNA/RNA UD Indexes Set D, Tagmentation (96 Indexes, 96 Samples)
20091660
Illumina® DNA/RNA UD Indexes Set D, Tagmentation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples. Purchase library prep and enrichment reagents and probe panels separately.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
Illumina 5-Base DNA Prep with Enrichment uses a one-step enzymatic process to convert 5mC to T, yielding greater nucleotide diversity than traditional bisulfite treatment. Optimized ligation-based library preparation is followed by target-capture based enrichment, with customized panel design. When paired with specialized analysis, the solution enables simultaneous detection of 5mC and genomic variants in a single assay.
Illumina 5-Base DNA Prep with Enrichment enables high-sensitivity methylation profiling and variant detection in one simple, scalable, and cost-effective workflow, with easy customization of target regions.
Illumina 5-Base DNA Prep with Enrichment requires a custom panel design and is compatible with Illumina Custom Enrichment Panel v2. Panels can be designed and ordered using DesignStudio software. The specialized design algorithm accommodates the 5mC-to-T base conversion for efficient capture of both methylated and nonmethylated targets. Panels made for methods, such as bisulfite sequencing, that convert unmethylated cytosines, are not compatible.
DRAGEN secondary analysis uses innovative 5-base algorithms to provide accurate annotation of both genome and methylome from a single dataset. An easy checkbox option activates methylation reporting features within the DRAGEN Enrichment pipeline.
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.