
NovaSeq X Series ordering
Advanced chemistry, optics, and informatics combine to deliver exceptional sequencing speed and data quality, outstanding throughput, and scalability.
A rapid, cost-effective workflow for accurate, unbiased detection of the protein-coding transcriptome with precise measurement of strand information.
Assay time
Hands-on time
Input quantity
Illumina Stranded mRNA Prep provides a streamlined RNA sequencing (RNA-Seq) solution for clear and comprehensive analysis across the coding transcriptome for discovery of features such as novel isoforms, gene fusions, and allele-specific expression.
*Not compatible with FFPE samples
| Assay time | 6.5 hr |
|---|---|
| Automation capability | Liquid handling robot(s) |
| Automation details | Explore available automation methods |
| Content specifications | Captures the coding transcriptome with strand information |
| Description | A simple, cost-effective solution for analysis of the coding transcriptome with precise strand information |
| Hands-on time | < 3 hr |
| Input quantity | 25-1000 ng standard-quality total RNA |
| Instruments | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System |
| Mechanism of action | PolyA capture, ligation-based addition of adapters and indexes |
| Method | mRNA sequencing |
| Multiplexing | Up to 384 Unique Dual Indexes (UDIs) |
| Nucleic acid type | RNA |
| Specialized sample types | Not FFPE-compatible, Low-input samples |
| Species category | Mammalian, Bovine, Mouse, Human, Rat |
| Species details | Works with high-quality RNA from any species with polyA tails |
| Strand specificity | Stranded |
| System compatibility details | Library prep is designed to be compatible with all Illumina sequencing systems, and is extensively validated on the NextSeq 500/550 and NovaSeq 6000 Systems. |
| Technology | Sequencing |
| Variant class | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants |
Choose the library prep kit size (16 sample or 96 sample) that meets your processing needs.
Unique dual indexes are sold separately and required for multiplexing. Choose the indexing option that best fits your target sequencing throughput. You can add Set A, B, C, and D together to sequence up to 384 samples in a single run. Sequencing throughput and multiplexing capabilities can vary depending on sequencer and flow cell reagents.
Illumina Stranded mRNA Prep enables preparation of sequencing-ready transcriptome libraries in a single day with low sample inputs of standard (non-degraded) RNA from a wide range of sample types.
Illumina Stranded mRNA Prep
| Instrument | Recommended number of samples | Read length |
|---|---|---|
| NextSeq 550 System | Mid output: 5 |
2 × 75 bp |
| NextSeq 2000 System | P2: 16 |
2 × 75 bp |
| NovaSeq 6000 System | SP: 32 |
2 × 75 bp |
mRNA sequencing (mRNA-Seq) using NGS can identify both known and novel transcripts, and measure transcript abundance.
Learn how to analyze transcriptome changes or profile genome-wide gene expression levels in a single experiment with next-generation sequencing methods.
Drug response RNA biomarker analysis
Assessment of RNA-based biomarkers predictive of therapeutic response is increasingly becoming an integral part of the drug development process.
| Illumina Stranded mRNA Prep | TruSeq Stranded mRNA | Illumina RNA Prep with Enrichment | Illumina Stranded Total RNA Prep with Ribo-Zero Plus or Ribo-Zero Plus Microbiome | |
|---|---|---|---|---|
| Assay time | 6.5 hr | ~10.5 hr | < 9 hr | ~7 hr |
| Automation capability | Liquid handling robot(s) | Liquid handling robot(s) | Liquid handling robot(s) | Liquid handling robot(s) |
| Automation details | Explore available automation methods | Explore available automation methods | ||
| Content specifications | Captures the coding transcriptome with strand information | Captures the coding transcriptome with strand information | Captures the coding transcriptome when used with Illumina Exome Panel | Captures coding RNA plus multiple forms of non-coding RNA |
| Description | A simple, cost-effective solution for analysis of the coding transcriptome with precise strand information | Gives researchers a clear, comprehensive view of the coding transcriptome with precise strand information. | A reproducible, economical solution enabling targeted transcript detection and discovery from a broad range of sample types and inputs including formalin-fixed, paraffin- embedded (FFPE) tissues and other low-quality samples | For the preparation of whole-transcriptome sequencing-ready libraries from low RNA inputs and low-quality samples that supports a wide range of RNA-Seq applications. Includes Ribo-Zero Plus for single-tube depletion of rRNA and globin RNA from multiple species to facilitate rich transcriptome analyses. |
| Hands-on time | < 3 hr | ~4.5 hr | < 2 hr | < 3 hr |
| Input quantity | 25-1000 ng standard-quality total RNA | 0.1 – 1 ug total RNA or 10 - 100 ng previously isolated mRNA (from species with polyA tails) | 10 ng RNA; 20 ng FFPE RNA | 1–1000 ng total RNA for standard-quality RNA samples. 10 ng total RNA minimum recommended for optimal performance and FFPE samples |
| Instruments | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System | MiSeq System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiniSeq System, NovaSeq 6000 System, MiSeq i100 System, MiSeq i100 Plus System | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System |
| Mechanism of action | PolyA capture, ligation-based addition of adapters and indexes | Oligo-dT beads capture polyA tails | Bead-linked transposome | Enzymatic rRNA depletion, ligation-based addition of adapters and indexes |
| Method | mRNA sequencing | mRNA sequencing | mRNA sequencing, Targeted RNA sequencing, Target enrichment | Whole-transcriptome sequencing, Metatranscriptome sequencing |
| Multiplexing | Up to 384 Unique Dual Indexes (UDIs) | 1–96 | Up to 384 Unique Dual Indexes (UDIs) | Up to 384 Unique Dual Indexes (UDIs) |
| Nucleic acid type | RNA | RNA | RNA | RNA |
| Specialized sample types | Not FFPE-compatible, Low-input samples | Not FFPE-compatible | Blood, Low-input samples, FFPE tissue | Microbiome samples, Low-input samples, FFPE tissue |
| Species category | Mammalian, Bovine, Mouse, Human, Rat | Mammalian, Bovine, Mouse, Human, Rat | Human, Virus | Mouse, Human, Rat, Bacteria |
| Species details | Works with high-quality RNA from any species with polyA tails | Works with high-quality RNA from any species with polyA tails | Ribo-Zero Plus depletes abundant transcripts from multiple species including: human cytoplasmic and mitochondrial rRNA, mouse rRNA, rat rRNA, bacteria (E. coli and B. subtilis) rRNA, and human beta globin transcripts Ribo-Zero Plus Microbiome depletes abundant transcripts from multiple species including: those depleted in Ribo-Zero Plus as well as diverse microbial species and ATCC MSA-2002, MSA-2005, and MSA-2006 | |
| Strand specificity | Stranded | Stranded | Non-stranded | Stranded |
| System compatibility details | Library prep is designed to be compatible with all Illumina sequencing systems, and is extensively validated on the NextSeq 500/550 and NovaSeq 6000 Systems. | |||
| Technology | Sequencing | Sequencing | Sequencing | Sequencing |
| Variant class | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants |
Library Prep & Array Kit Selector
Find the right sequencing library preparation kit or microarray for your needs. Filter by method, species, and more. Compare, share, and order kits.
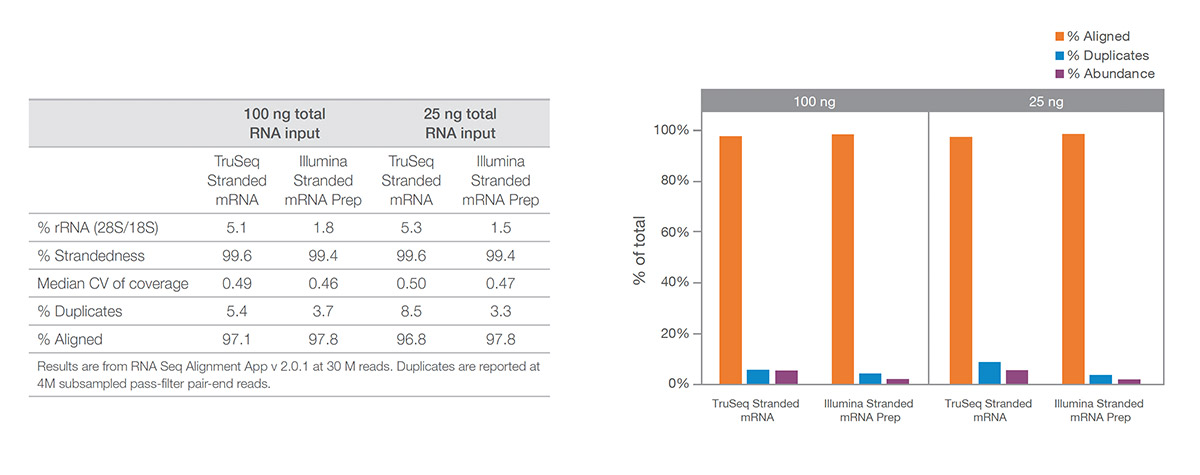

(Left) Performance metrics for Illumina Stranded mRNA Prep. (Right) Illumina Stranded mRNA Prep was compared against TruSeq Stranded mRNA. Illumina Stranded mRNA Prep showed superior performance, particularly with an input of 25 ng UHR RNA.
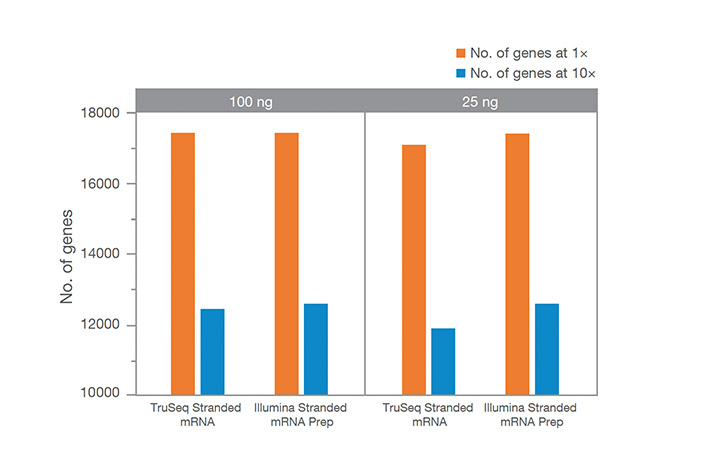

Illumina Stranded mRNA Prep enables greater gene detection with low RNA inputs, as compared to TruSeq Stranded mRNA. The number of genes detected is reported at 30M subsampled paired-end reads PF. More genes detected at 1× coverage is an indicator of greater sensitivity.
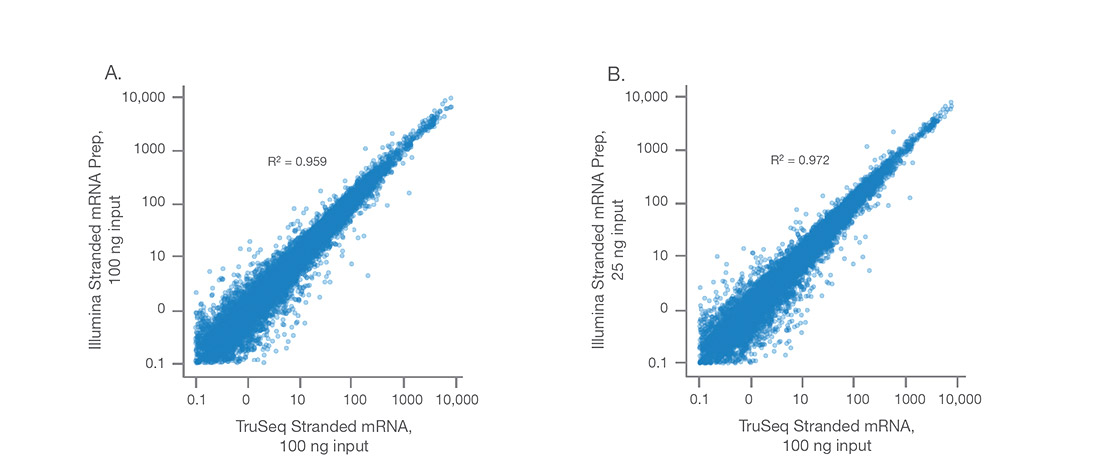

Illumina Stranded mRNA Prep produces highly concordant data with TruSeq Stranded mRNA at (A) equivalent inputs of 100 ng UHR RNA and (B) with reduced input of 25 ng vs 100 ng UHR RNA.
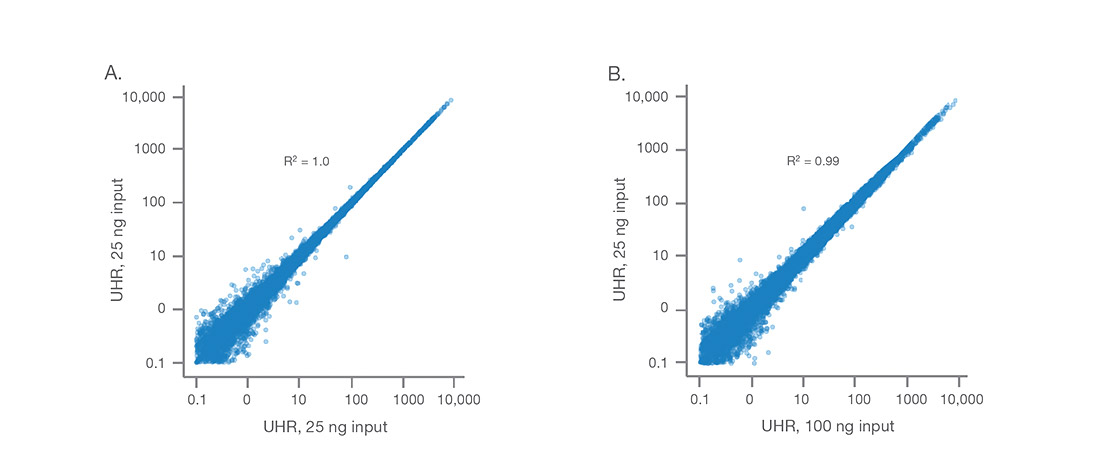

Illumina Stranded mRNA Prep achieves high data concordance between (A) technical replicates of 25 ng UHR RNA and (B) between input amounts of 25 ng and 100 ng UHR RNA. Libraries were sequenced at 2 × 74 bp, subsampled to 30M reads. Data analysis was performed using the BaseSpace RNA-Seq Alignment App.
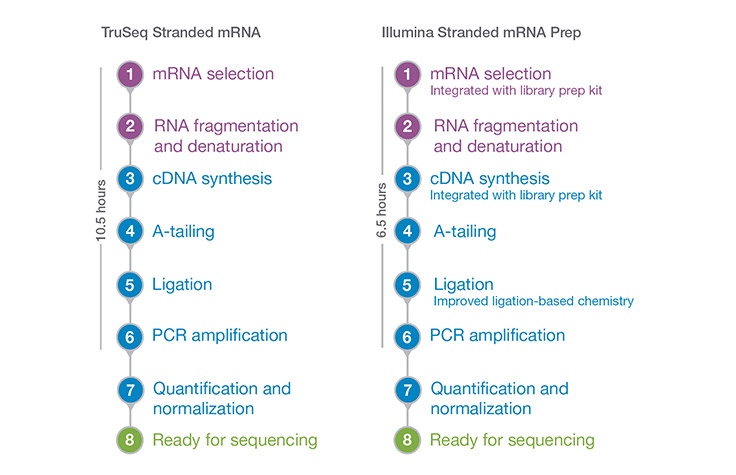

Illumina Stranded mRNA Prep delivers a fast workflow with reduced hands-on time. Times may vary depending on equipment used, number of samples processed, automation procedures, or user experience.
To accurately determine gene expression from overlapping genes, it is important to retain the information found on the strand of origin. Stranded RNA-Seq allows the first and second cDNA strands to be distinguished so that the second strand can be degraded while the first cDNA strand (strand of origin) will undergo further PCR amplification.
Library preparation takes 6.5 hr with less than 3 hr of hands-on time.
Oligo(dT) magnetic beads capture and purify the mRNA containing polyA tails. The purified mRNA is fragmented and copied into first-strand cDNA using reverse transcriptase and random primers. In a second-strand cDNA synthesis step, dUTP replaces dTTP to achieve strand specificity. The final steps add adenine and thymine bases to fragment ends and ligate adapters. The resulting products are purified and selectively amplified for sequencing.
Illumina® Stranded mRNA Prep, Ligation (16 Samples).
20040532
A simple, scalable, cost-effective, rapid single-day solution for analyzing the coding transcriptome leveraging as little as 25 ng of total RNA input. Kit contains library prep reagents for 16 samples. Indexes sold separately, compatible only with IDT for Illumina RNA UD indexes, Ligation.
List Price:
Discounts:
Illumina® Stranded mRNA Prep, Ligation (96 Samples).
20040534
A simple, scalable, cost-effective, rapid single-day solution for analyzing the coding transcriptome leveraging as little as 25 ng of total RNA input. Kit contains library prep reagents for 96 samples. Indexes sold separately, compatible only with IDT for Illumina RNA UD indexes, Ligation.
List Price:
Discounts:
Illumina® RNA UD Indexes Set A, Ligation (96 Indexes, 96 Samples)
20091655
Illumina® RNA UD Indexes Set A, Ligation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples and RNA Index Anchor plate. Used with Illumina Stranded Total RNA Prep and Illumina Stranded mRNA Prep Kits. Purchase library prep reagents separately.
List Price:
Discounts:
Illumina® RNA UD Indexes Set B, Ligation (96 Indexes, 96 Samples)
20091657
Illumina® RNA UD Indexes Set B, Ligation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples and RNA Index Anchor plate. Used with Illumina Stranded Total RNA Prep and Illumina Stranded mRNA Prep Kits. Purchase library prep reagents separately.
List Price:
Discounts:
Illumina® RNA UD Indexes Set C, Ligation (96 Indexes, 96 Samples)
20091659
Illumina® RNA UD Indexes Set C, Ligation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples and RNA Index Anchor plate. Used with Illumina Stranded Total RNA Prep and Illumina Stranded mRNA Prep Kits. Purchase library prep reagents separately.
List Price:
Discounts:
Illumina® RNA UD Indexes Set D, Ligation (96 Indexes, 96 Samples)
20091661
Illumina® RNA UD Indexes Set D, Ligation (96 Indexes, 96 Samples) Includes 96, 10 bp indexes sufficient for labeling 96 samples and RNA Index Anchor plate. Used with Illumina Stranded Total RNA Prep and Illumina Stranded mRNA Prep Kits. Purchase library prep reagents separately.
List Price:
Discounts:
Illumina Stranded mRNA LP Kit Training - Customer Site
20044765
2-day on-site training for the Illumina Stranded mRNA Library Preparation Kit. Hands-on training includes quantification of mRNA input as well as best practices, troubleshooting tips, and training on Illumina-supported analysis tools specific to the application workflow.
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.